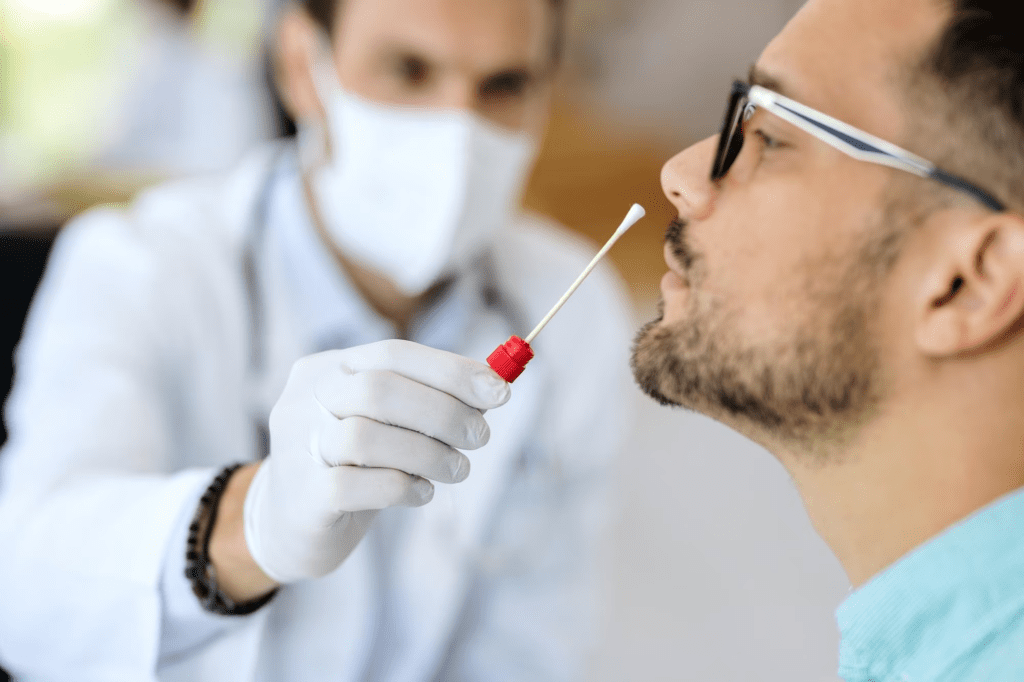
PCR
For standard PCR, all you need is a DNA polymerase, magnesium, nucleotides, primers, the DNA template to be amplified and a thermocycler. The PCR mechanism is as simple as its purpose: 1) double-stranded DNA (dsDNA) is heat denatured, 2) primers align to the single DNA strands and 3) the primers are extended by DNA polymerase, resulting in two copies of the original DNA strand. The denaturation, annealing, and elongation process over a series of temperatures and times is known as one cycle of amplification. Each step of the cycle should be optimized for the template and primer set used. This cycle is repeated approximately 20-40 times and the amplified product can then be analyzed. PCR is widely used to amplify DNA for subsequent experimental use. PCR also has applications in genetic testing or for the detection of pathogenic DNA.

As PCR is a highly sensitive method and very small volumes are required for single reactions, preparation of a master mix for several reactions is recommended. The master mix must be well mixed and then split by the number of reactions, ensuring that each reaction will contain the same amount of enzyme, dNTPs and primers. Many suppliers, such as Enzo Life Sciences, also offer PCR mixes that already contain everything except primers and the DNA template.
Guanine/Cytosine-rich (GC-rich) regions represent a challenge in standard PCR techniques. GC-rich sequences are more stable than sequences with lower GC content. Furthermore, GC-rich sequences tend to form secondary structures, such as hairpin loops. As a result, GC-rich double strands are difficult to completely separate during the denaturation phase. Consequently, DNA polymerase cannot synthesize the new strand without hindrance. A higher denaturation temperature can improve this and adjustments towards a higher annealing temperature and shorter annealing time can prevent unspecific binding of GC-rich primers. Additional reagents can improve the amplification of GC-rich sequences. DMSO, glycerol and betaine help to disrupt the secondary structures that are caused by GC interactions and thereby facilitate separation of the double strands.
RT-PCR
Reverse transcription PCR, or RT-PCR, allows the use of RNA as a template. An additional step allows the detection and amplification of RNA. The RNA is reverse transcribed into complementary DNA (cDNA), using reverse transcriptase. The quality and purity of the RNA template is essential for the success of RT-PCR. The first step of RT-PCR is the synthesis of a DNA/RNA hybrid. Reverse transcriptase also has an RNase H function, which degrades the RNA portion of the hybrid. The single stranded DNA molecule is then completed by the DNA-dependent DNA polymerase activity of the reverse transcriptase into cDNA. The efficiency of the first-strand reaction can affect the amplification process. From here on, the standard PCR procedure is used to amplify the cDNA. The possibility to revert RNA into cDNA by RT-PCR has many advantages. RNA is single-stranded and very unstable, which makes it difficult to work with. Most commonly, it serves as a first step in qPCR, which quantifies RNA transcripts in a biological sample.
tag : RT-PCT TEST , RT PCR PHUKET , PATONG PCR TEST , PHUKET PCR TEST , Phuket Cheap Covid Test , RT-PCR Cheap Test , RT PCR Promotion , PCR Test Near Me , PCR Kata , PCR Paton , PCR-Test Karon , RT-PCR Karon Beach Paton, Sainamyen, Sai Nam Yen, Kata, 2023, New Year